Process microorganism genetic data
obtained by NGS and analyze it
using a complete and intuitive toolkit

- We have been operating in the market since 2021.
- More than 4 thousand samples processed per year.
- More than 7 thousand analyses conducted
- More than 17 thousand different organisms identified.
Meet Varsmetagen pipelines
Virome
The virome is a type of metagenomic analysis performed on sequencing data from clinical or environmental samples, with the aim of identifying and characterizing viral agents present in the sample. With Varsmetagen, it is possible to inspect quality metrics, identify viral pathogens, obtain graphical representations of the abundance and diversity of the microbiota (viruses, bacteria, and fungi), recover complete viral genomes, and perform paired analyses for reporting and report generation.
Microbiome
The virome is a type of metagenomic analysis performed on sequencing data from clinical or environmental samples, with the aim of identifying and characterizing viral agents present in the sample. With Varsmetagen, it is possible to inspect quality metrics, identify viral pathogens, obtain graphical representations of the abundance and diversity of the microbiota (viruses, bacteria, and fungi), recover complete viral genomes, and perform paired analyses for reporting and report generation.
Metagenomics
The bioinformatics pipeline for metagenomes enables the detection of various pathogens (bacteria, viruses, fungi, and eukaryotic parasites), as well as symbiotic microorganisms, through taxonomic analysis of reads and contigs, recovery of metagenome-assembled genomes (MAGs), and targeted mapping to a reference genome. With Varsmetagen, it is possible to perform the diagnosis of infectious diseases, characterize the microbiome and mycobiome, and generate comprehensive reports and diagnoses based on the results.
Viroma
Viroma é um tipo de análise de metagenômica realizada em dados de sequenciamento de amostras clínicas ou ambientais, onde o objetivo é a identificação e caracterização dos agentes virais presentes na amostra. Com o Varsmetagen, é possível inspecionar métricas de qualidade, identificar patógenos virais, obter representações gráficas de abundância e diversidade da microbiota (vírus, bactérias e fungos), recuperar genomas virais completos e realizar análises pareadas para laudamento e geração de relatórios.
Microbioma
O pipeline de Microbioma (Metataxonomia ou Metabarcoding) oferece um tipo de análise para Amplicon (16S e ITS) realizada em dados de sequenciamento de amostras clínicas ou ambientais, onde o objetivo é avaliar a diversidade de microrganismos e identificação de disbioses presente na amostra. Com o Varsmetagen, é possível comparar os resultados com o banco de referência do Human Microbiome Project (HMP), além de inspecionar métricas de qualidade, observar disbioses e obter representações gráficas de abundância e diversidade do microbioma (bactérias e fungos).
Metagenômica
O pipeline de bioinformática para metagenomas possibilita a detecção de diversos patógenos (bactérias, vírus, fungos e parasitas eucarióticos) e também de microrganismos simbióticos, por meio de análises taxonômicas de reads e contigs, recuperação de genomas de metagenomas (MAGs) e mapeamento direcionado em genoma de referência. Com o Varsmetagen é possível realizar o diagnóstico de doenças infecciosas, caracterizar o microbioma e micobioma, e emitir laudos e relatórios completos sobre os resultados.
Varsmetagen features
Our cloud-based software allows researchers and healthcare professionals to investigate microbial genetic data in a fast, efficient, and scalable way.
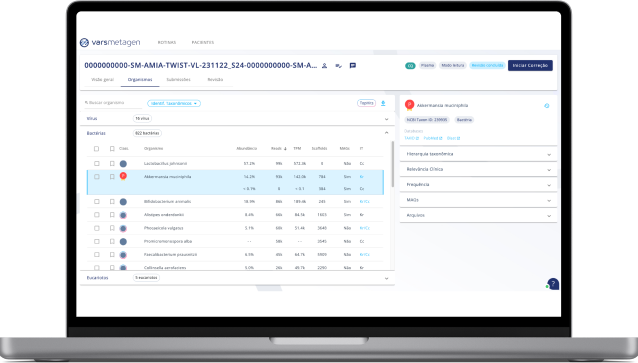
1. Graphic tools
Use pie charts, sunburst charts, and useful tables to get the best out of your data.

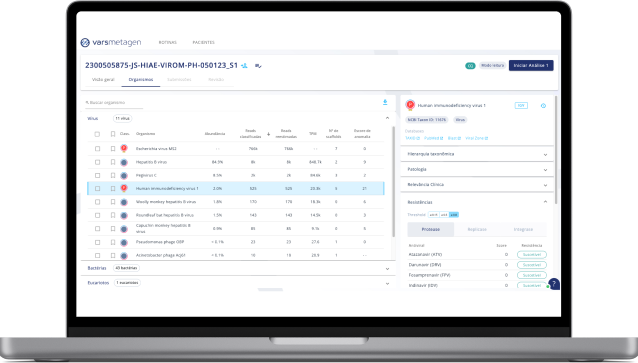
2. Anomaly meter
Artificial Intelligence algorithms developed in a unique way for providing anomaly scores to our users, allowing them to distinguish normal microbiota species from those acting as pathogens.

3. Intuitive Interface
Our state-of-the-art tools were developed minding different types of users. Our clients don’t need advanced coding knowledge to perform analysis but advanced features are available.

4. Security
As the best hospital in Latin America, Hospital Israelita Albert Einstein and Varsmetagen uses the best industry protocols to keep your and your patients data safe. All data submitted to our platform are protected by our strict privacy policy.

Success Cases
SARS-Omics Project
The SARS-Omics Project emerged at the beginning of the COVID-19 pandemic and performs the sequencing of the COVID-19 virus, SARS-CoV-2, as well as studying co-infections with other viruses, bacteria, and fungi in the respiratory tract of COVID-19 patients. A tremendous amount of NGS data has been generated by the project, and the practicality and scalability of Varsmetagen have been helpful in the process of analyzing and interpreting these results.

Scientific Production
João N. de Almeida Jr, André Mario Doi, Maria Julia L. Watanabe, Maira Maraghello Maluf, Cecília Leon Calderon, Moacyr Silva Jr, Jacyr Pasternak, Paula Célia M. Koga, Kelly Aline S. Santiago, Luis Fernando C. Aranha, Gilberto Szarf, Gustavo B. da Silva Teles, Renée Zon Filippi, Vitor Ribeiro Paes, Marina Baeta, Nelson Hamerschlak, Cristovão Luis P. Mangueira, Marines Dalla Valle Martino
DOI
10.1111/
myc.13433
DOI
10.1136/
pn-2023-003795
DOI
10.1016/
j.spinee.2021.09.005
Fernanda de Mello Malta, Deyvid Amgarten, Ana Catharina de Seixas Santos Nastri, Yeh-Li Ho et al.
DOI
10.3201/
eid2606.200099
Roberta Cardoso Petroni, Anelisie da Silva Santos, Marcio Anunciação Menezes, Ana Paula Moreira Salles et al.
DOI
10.1016/
j.bjid.2021.102199
Marina Barrionuevo Mathias, Fernando Gatti Menezes, Gustavo Bruniera Peres Fernandes, Vitor Ribeiro Paes et al.
DOI
10.1212/
CPJ.0000000000200167
Lia Cunha, Adriana Luchs, Lais S. Azevedo, Vanessa C. M. Silva et al.
DOI
10.3390/
v15020335
Early users program




AI integrated into a metagenomic datalake
Varsmetagen is a platform based on the most advanced cloud computing technologies, which allows the creation of a metagenomic datalake for integrated analysis of omics data from institutions that wish to benefit from this technology. Through the datalake, it is possible to structure data from different types and sources, producing tables and repositories with centralized data for statistical analysis, training of artificial intelligence models, and dashboard visualizations. Many of the analyses enabled by the datalake have become features available to all organizations using Varsmetagen, such as the Anomaly meter™ and Varsdata™.
Meet our team of bioinformaticians
Deyvid Amgarten, PhD
Metagenomics / Viral genomics
Ana Carolina Soares, PhD
Metagenomics / Microbiology
Raquel
Castillo, PhD
Metagenomics
Raquel
Riyuzo, PhD
Metagenomics / Bacterial genomics
Erick
Dorlass, MSc
Viral genomics
Tania Girão Mangolini
Bioinformatics
Pedro
Sebe
Data science

Who are we?
Varsomics belongs to the clinical laboratory of Einstein. We are experts in bioinformatics with services applied to clinical diagnostics and academic research. Our main solutions are Varstation, a platform for processing human genetic variants, and Varsmetagen, a platform for identifying microorganisms from sequencing data.
Our mission:
To enable and promote access to the use of new genetic technologies to enhance medical treatments and population health.