Customized analysis from genetic data
Process and analyze human genetic data generated from Next-Generation Sequencing (NGS).
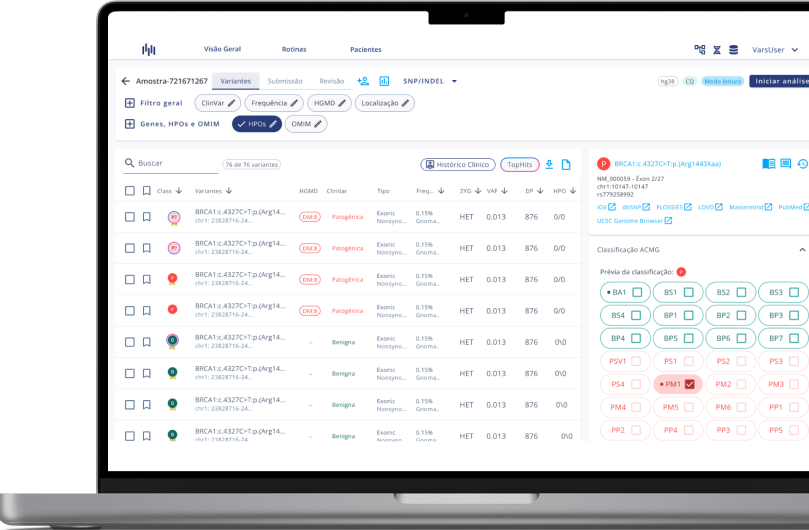
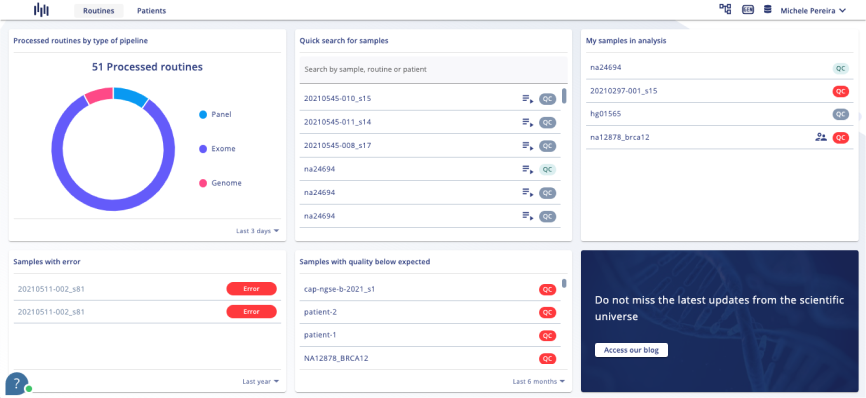

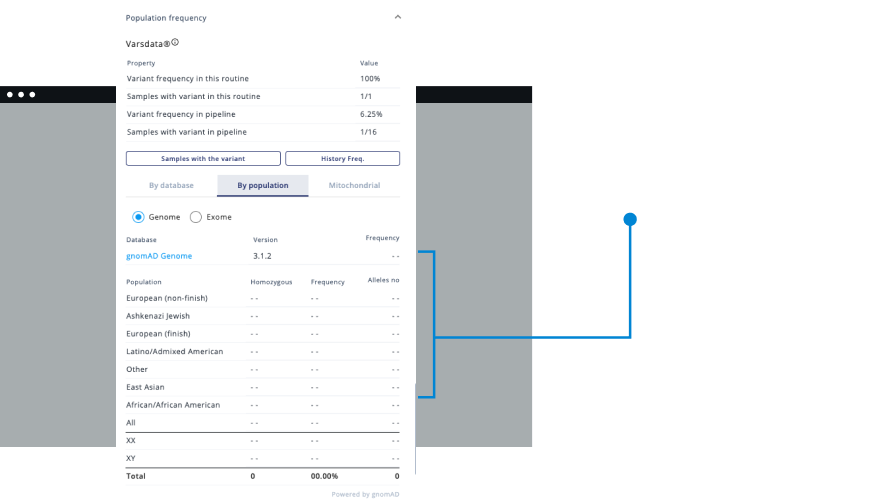
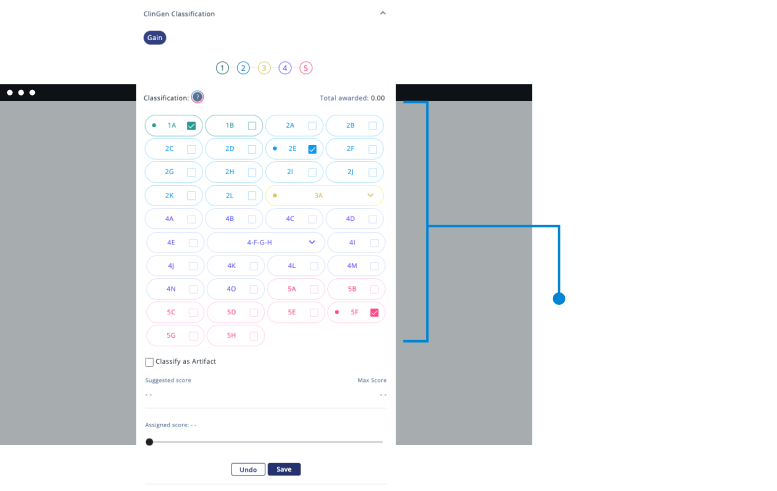
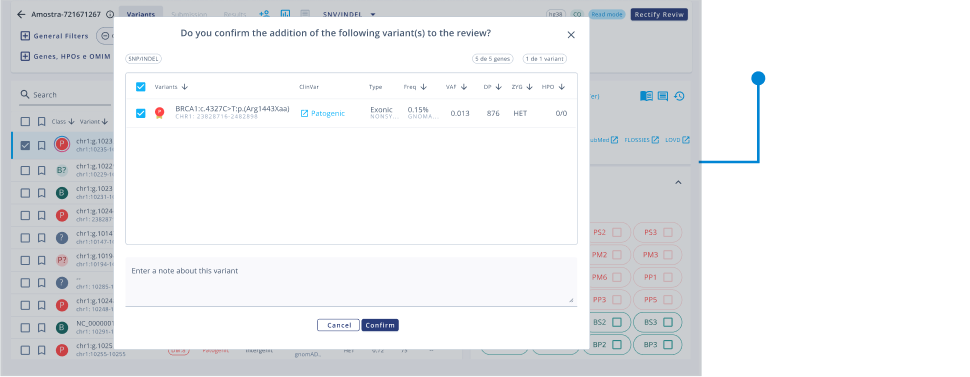
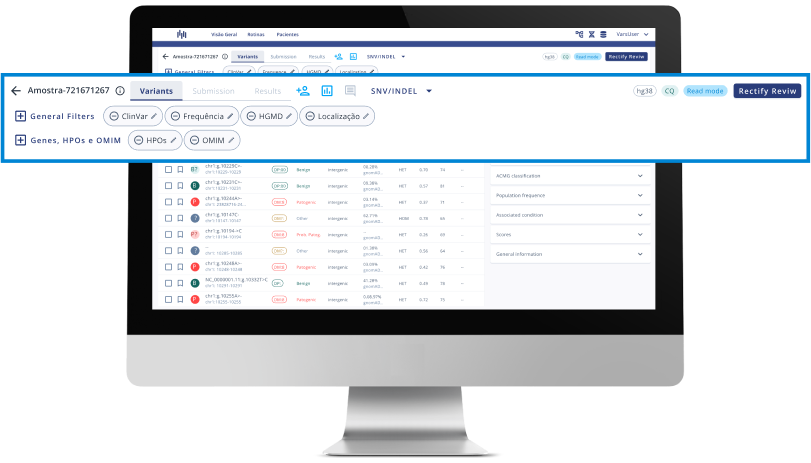
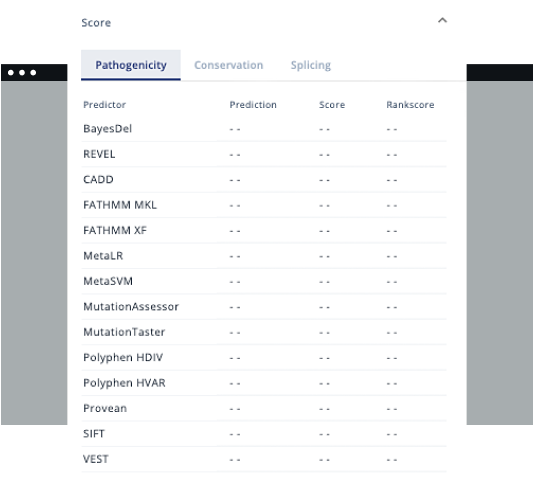
- Carry out analysis following College of American Pathologists guidelines.
- Curated clinically relevant variants, making data easier to be interpreted.
- Dynamic and custom filters to create and save pipelines.
Ask your demonstration
for Varstation 3.0®
Ask your demonstration
for Varstation 3.0®
- Carry out analysis following following College of American Pathologists guidelines.
- Curated clinically relevant variants, making data easier to be interpreted.
- Dynamic and custom filters to create and save pipelines.
How does it work?
Set your pipeline
Inform your sequencing platform, library preparation technique and analysis type to configure your personalized pipelines.
Sample upload
Upload from a local drive or directly from Illumina’s Basespace. We support .FASTQ; .BAM and .VCFs
Curation of clinically relevant variants
Genetic variants are automatically pre-classified by a proper algorithm, and the filter system delimits variants that may be related to clinical suspicions.
Review and final report
Review the findings according to the quality recommendations of the accreditation agencies and generate an analysis report with the final result.
Advantages of using Varstation®
Analysis time optimization
Significantly reduce and optimize the time for processing, sorting variants and generating reports from NGS human data.
End-to-end automated processing
Quality parameter evaluation, mapping, multi-variant callers, database annotation, and automatic pre-sorting following ACMG, AMP, PALC, and CAP guidelines.
Clinical interpretation support
Varstation grants you easy access to more than 200 genetic variant databases. Those include data for germline and somatic analysis.
Data filtering
Data filtering mechanism that incorporates all annotated mutation data - including human phenotypes, patient clinical history and associated diseases (OMIM/UniProt).

We are part of the best Hospital in Latin America:
We preserve the diversity, quality and organizational culture of Hospital Israelita Albert Einstein, considered by the América Economía Intelligence’s ranking as the best hospital in Latin America.
Trusted by










Affordable prices per processing
Flexibility to enable fast changes in your analytical processes and a powerful pay-per-use cloud solution.
Sucess Case
During the COVID-19 pandemic, Varsomics, in partnership with Albert Einstein Israelite Hospital, developed the world’s first test for the diagnosis of the new coronavirus based on Next-Generation Sequencing (NGS).
The test, which was known as VarsVid, identifies the presence of the virus, functioning as a diagnostic tool to be used from the first day of infection, in the same way as RT-PCR, which makes it an alternative for adopting a diagnostic test.
Processing takes up to 72 hours, and has 95% sensitivity and 99% specificity, or positive, there are no cases of false.